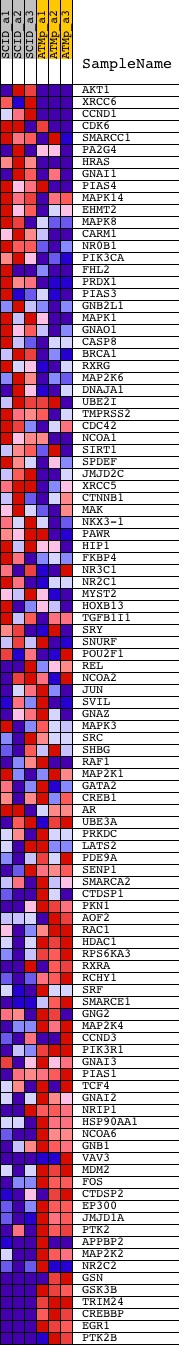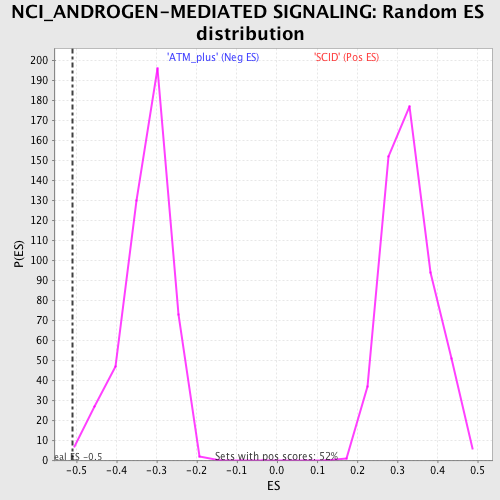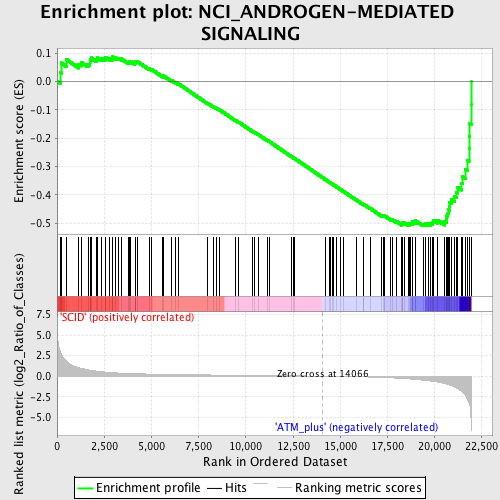
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NCI_ANDROGEN-MEDIATED SIGNALING |
| Enrichment Score (ES) | -0.5102 |
| Normalized Enrichment Score (NES) | -1.5689354 |
| Nominal p-value | 0.004149378 |
| FDR q-value | 0.3274345 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 182 | 2.933 | 0.0309 | No | ||
| 2 | XRCC6 | 1417437_at 1428504_at 1442250_at | 221 | 2.685 | 0.0651 | No | ||
| 3 | CCND1 | 1417419_at 1417420_at 1448698_at | 504 | 1.821 | 0.0766 | No | ||
| 4 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 1148 | 1.022 | 0.0609 | No | ||
| 5 | SMARCC1 | 1423416_at 1423417_at 1455246_at 1459824_at | 1291 | 0.939 | 0.0669 | No | ||
| 6 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 1643 | 0.770 | 0.0612 | No | ||
| 7 | HRAS | 1422407_s_at 1424132_at | 1741 | 0.725 | 0.0664 | No | ||
| 8 | GNAI1 | 1427510_at 1434440_at 1454959_s_at | 1743 | 0.724 | 0.0761 | No | ||
| 9 | PIAS4 | 1418861_at 1455394_at | 1800 | 0.703 | 0.0829 | No | ||
| 10 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 2079 | 0.612 | 0.0784 | No | ||
| 11 | EHMT2 | 1426888_at 1460692_at | 2155 | 0.591 | 0.0829 | No | ||
| 12 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 2363 | 0.538 | 0.0806 | No | ||
| 13 | CARM1 | 1419743_s_at 1442245_at | 2540 | 0.500 | 0.0792 | No | ||
| 14 | NR0B1 | 1417760_at | 2574 | 0.492 | 0.0843 | No | ||
| 15 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 2775 | 0.456 | 0.0813 | No | ||
| 16 | FHL2 | 1419184_a_at | 2910 | 0.432 | 0.0809 | No | ||
| 17 | PRDX1 | 1416000_a_at 1433866_x_at 1434731_x_at 1436691_x_at | 2912 | 0.432 | 0.0866 | No | ||
| 18 | PIAS3 | 1421646_a_at 1451115_at | 3115 | 0.400 | 0.0827 | No | ||
| 19 | GNB2L1 | 1440120_at 1455168_a_at | 3243 | 0.383 | 0.0820 | No | ||
| 20 | MAPK1 | 1419568_at 1426585_s_at 1442876_at 1453104_at | 3406 | 0.363 | 0.0795 | No | ||
| 21 | GNAO1 | 1421152_a_at 1448031_at 1460383_at | 3759 | 0.328 | 0.0678 | No | ||
| 22 | CASP8 | 1424552_at | 3828 | 0.322 | 0.0690 | No | ||
| 23 | BRCA1 | 1424629_at 1424630_a_at 1427698_at 1443763_at 1451417_at | 3894 | 0.317 | 0.0702 | No | ||
| 24 | RXRG | 1418782_at | 4125 | 0.299 | 0.0637 | No | ||
| 25 | MAP2K6 | 1426850_a_at 1459292_at | 4140 | 0.298 | 0.0670 | No | ||
| 26 | DNAJA1 | 1416288_at 1437220_x_at 1445729_at 1459835_s_at 1460179_at | 4158 | 0.297 | 0.0703 | No | ||
| 27 | UBE2I | 1422712_a_at 1422713_a_at 1425478_x_at 1453189_at | 4263 | 0.291 | 0.0694 | No | ||
| 28 | TMPRSS2 | 1419154_at 1449369_at 1458347_s_at 1459510_at | 4884 | 0.258 | 0.0444 | No | ||
| 29 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 5007 | 0.252 | 0.0422 | No | ||
| 30 | NCOA1 | 1418594_a_at 1434515_at 1439883_at 1443397_at 1443438_at 1459277_at | 5594 | 0.227 | 0.0184 | No | ||
| 31 | SIRT1 | 1418640_at 1458538_at | 5628 | 0.226 | 0.0200 | No | ||
| 32 | SPDEF | 1450220_a_at | 6077 | 0.209 | 0.0022 | No | ||
| 33 | JMJD2C | 1424458_at 1430628_at 1447091_at | 6254 | 0.203 | -0.0031 | No | ||
| 34 | XRCC5 | 1451968_at | 6415 | 0.198 | -0.0078 | No | ||
| 35 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 7984 | 0.149 | -0.0776 | No | ||
| 36 | MAK | 1422724_at 1422725_at | 8271 | 0.142 | -0.0888 | No | ||
| 37 | NKX3-1 | 1449998_at | 8435 | 0.138 | -0.0944 | No | ||
| 38 | PAWR | 1444808_at 1445390_at | 8608 | 0.134 | -0.1005 | No | ||
| 39 | HIP1 | 1424755_at 1424756_at 1432017_at 1432230_at 1434557_at 1444873_at | 9448 | 0.113 | -0.1374 | No | ||
| 40 | FKBP4 | 1416362_a_at 1416363_at 1458729_at 1459808_at | 9617 | 0.109 | -0.1436 | No | ||
| 41 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 10372 | 0.091 | -0.1770 | No | ||
| 42 | NR2C1 | 1418605_at 1439490_at 1446194_at 1449157_at 1458338_x_at | 10442 | 0.090 | -0.1789 | No | ||
| 43 | MYST2 | 1433433_at 1440746_at 1447631_at | 10665 | 0.085 | -0.1880 | No | ||
| 44 | HOXB13 | 1419576_at | 11124 | 0.074 | -0.2079 | No | ||
| 45 | TGFB1I1 | 1418136_at | 11253 | 0.071 | -0.2129 | No | ||
| 46 | SRY | 1450578_at 1450579_x_at | 12404 | 0.043 | -0.2650 | No | ||
| 47 | SNURF | 1459604_at | 12511 | 0.041 | -0.2693 | No | ||
| 48 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 12594 | 0.039 | -0.2725 | No | ||
| 49 | REL | 1420710_at | 14232 | -0.005 | -0.3474 | No | ||
| 50 | NCOA2 | 1435233_at 1435234_at 1446827_at 1450458_at | 14408 | -0.011 | -0.3553 | No | ||
| 51 | JUN | 1417409_at 1448694_at | 14497 | -0.014 | -0.3591 | No | ||
| 52 | SVIL | 1427662_at 1460694_s_at 1460735_at | 14581 | -0.016 | -0.3627 | No | ||
| 53 | GNAZ | 1426517_at 1435268_at | 14658 | -0.019 | -0.3660 | No | ||
| 54 | MAPK3 | 1427060_at | 14783 | -0.022 | -0.3713 | No | ||
| 55 | SRC | 1423240_at 1450918_s_at | 14992 | -0.030 | -0.3805 | No | ||
| 56 | SHBG | 1421602_at | 15151 | -0.035 | -0.3872 | No | ||
| 57 | RAF1 | 1416078_s_at 1425419_a_at | 15837 | -0.063 | -0.4178 | No | ||
| 58 | MAP2K1 | 1416351_at | 16232 | -0.082 | -0.4347 | No | ||
| 59 | GATA2 | 1428816_a_at 1450333_a_at | 16243 | -0.082 | -0.4341 | No | ||
| 60 | CREB1 | 1421582_a_at 1421583_at 1423402_at 1428755_at 1452529_a_at 1452901_at | 16598 | -0.105 | -0.4489 | No | ||
| 61 | AR | 1422982_at 1437064_at 1446340_at 1455647_at | 17182 | -0.150 | -0.4736 | No | ||
| 62 | UBE3A | 1416680_at 1416681_at 1416682_at 1425206_a_at 1431224_at 1445727_at | 17278 | -0.159 | -0.4758 | No | ||
| 63 | PRKDC | 1451576_at | 17327 | -0.164 | -0.4758 | No | ||
| 64 | LATS2 | 1419678_at 1419679_at 1425420_s_at 1426156_at 1437101_at 1439441_x_at | 17351 | -0.166 | -0.4746 | No | ||
| 65 | PDE9A | 1449403_at | 17676 | -0.199 | -0.4868 | No | ||
| 66 | SENP1 | 1424330_at 1428925_at 1451319_at | 17773 | -0.210 | -0.4884 | No | ||
| 67 | SMARCA2 | 1430526_a_at 1439930_at 1452333_at | 17965 | -0.232 | -0.4940 | No | ||
| 68 | CTDSP1 | 1452055_at | 18261 | -0.276 | -0.5038 | Yes | ||
| 69 | PKN1 | 1458674_at 1459231_at | 18267 | -0.277 | -0.5004 | Yes | ||
| 70 | AOF2 | 1426761_at 1426762_s_at | 18285 | -0.280 | -0.4974 | Yes | ||
| 71 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 18420 | -0.297 | -0.4996 | Yes | ||
| 72 | HDAC1 | 1448246_at | 18588 | -0.320 | -0.5029 | Yes | ||
| 73 | RPS6KA3 | 1427299_at 1441360_at 1452383_at | 18684 | -0.334 | -0.5028 | Yes | ||
| 74 | RXRA | 1425762_a_at 1430497_at 1454773_at | 18724 | -0.341 | -0.5000 | Yes | ||
| 75 | RCHY1 | 1432144_a_at | 18806 | -0.355 | -0.4990 | Yes | ||
| 76 | SRF | 1418255_s_at 1418256_at | 18829 | -0.361 | -0.4952 | Yes | ||
| 77 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 18964 | -0.387 | -0.4961 | Yes | ||
| 78 | GNG2 | 1418451_at 1418452_at 1428156_at 1428157_at | 18983 | -0.392 | -0.4917 | Yes | ||
| 79 | MAP2K4 | 1426233_at 1442679_at 1451982_at | 19388 | -0.482 | -0.5037 | Yes | ||
| 80 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 19489 | -0.507 | -0.5015 | Yes | ||
| 81 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 19644 | -0.549 | -0.5012 | Yes | ||
| 82 | GNAI3 | 1428645_at 1437225_x_at | 19794 | -0.592 | -0.5001 | Yes | ||
| 83 | PIAS1 | 1422581_at 1446448_at 1455486_at 1455611_at | 19898 | -0.621 | -0.4965 | Yes | ||
| 84 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 19931 | -0.634 | -0.4895 | Yes | ||
| 85 | GNAI2 | 1419449_a_at 1435652_a_at | 20160 | -0.718 | -0.4903 | Yes | ||
| 86 | NRIP1 | 1418469_at 1432603_at 1434384_at 1446389_at 1447211_at 1449089_at 1454295_at | 20522 | -0.881 | -0.4951 | Yes | ||
| 87 | HSP90AA1 | 1426645_at 1437497_a_at 1438902_a_at 1455078_at | 20613 | -0.930 | -0.4867 | Yes | ||
| 88 | NCOA6 | 1423374_at | 20631 | -0.939 | -0.4750 | Yes | ||
| 89 | GNB1 | 1417432_a_at 1425908_at 1454696_at 1459442_at | 20700 | -0.993 | -0.4648 | Yes | ||
| 90 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20714 | -1.006 | -0.4519 | Yes | ||
| 91 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 20769 | -1.045 | -0.4404 | Yes | ||
| 92 | FOS | 1423100_at | 20795 | -1.059 | -0.4273 | Yes | ||
| 93 | CTDSP2 | 1423660_at 1423661_s_at 1451075_s_at | 20875 | -1.119 | -0.4160 | Yes | ||
| 94 | EP300 | 1434765_at | 21052 | -1.301 | -0.4066 | Yes | ||
| 95 | JMJD1A | 1426810_at | 21136 | -1.403 | -0.3916 | Yes | ||
| 96 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 21220 | -1.521 | -0.3751 | Yes | ||
| 97 | APPBP2 | 1430265_at 1434039_at 1434040_at 1434041_at 1444886_at 1447855_x_at 1451251_at | 21402 | -1.799 | -0.3593 | Yes | ||
| 98 | MAP2K2 | 1415974_at 1443436_at 1460636_at | 21462 | -1.936 | -0.3360 | Yes | ||
| 99 | NR2C2 | 1425014_at 1439696_at 1445613_at 1446955_at 1451569_at 1454167_at 1454851_at | 21642 | -2.428 | -0.3117 | Yes | ||
| 100 | GSN | 1415812_at 1436991_x_at 1437171_x_at 1441225_at 1456312_x_at 1456568_at 1456569_x_at | 21737 | -2.841 | -0.2780 | Yes | ||
| 101 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21817 | -3.254 | -0.2380 | Yes | ||
| 102 | TRIM24 | 1427258_at 1427259_at 1446295_at 1459608_at | 21823 | -3.299 | -0.1941 | Yes | ||
| 103 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21856 | -3.508 | -0.1486 | Yes | ||
| 104 | EGR1 | 1417065_at | 21928 | -5.313 | -0.0807 | Yes | ||
| 105 | PTK2B | 1434653_at 1442437_at 1442927_at | 21933 | -6.050 | 0.0001 | Yes |